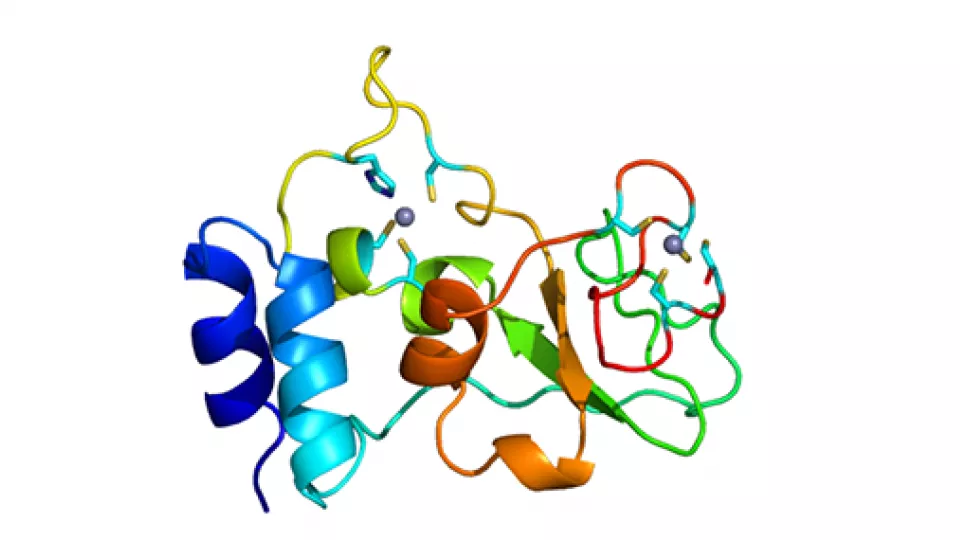
Recently we managed to solve the structure of one of these proteins, Nsp10, by using the BioMAX beamline at MAX IV Laboratory. Nsp10 is a critical component of the corona virus replication machinery.
Read more about our work in Swedish on the Lund University website, and in English on the websites of MAXIV, ESS, UCL and the League of advanced European Neutron Sources (LENS). The relevant data have been shared with the research community at the Covid-19 data portal Sweden. Our deposited data (PDB:6ZCT) is listed in the protein data module.
A story covering this collaborative project was broadcast on the Swedish news show TV4 Nyheterna.