LP3 Annual Report 2023
The “new” LP3 was created as a center in June 2016 by the Faculty of Science, the Faculty of Medicine and the Faculty of Engineering (LTH) by combining the “old” LP3, specialized in protein production, with LU’s protein crystallization facility. As part of the research infrastructure Protein Production Sweden (PPS), parts of LP3 are hosting and staffing the PPS Lund University node.
LP3’s part in PPS (see PPS homepage) is described in the PPS annual report to the Swedish research council. This report here will mainly focus on LP3 activities outside of PPS and try to highlight LP3’s local role for Lund University.
Outside PPS, LP3 continued to offer crystallization and protein crystal screening at the BioMAX beamline at MAX IV laboratory to its users. This enables non-experts in protein crystallography to use X-ray crystallography at MAX IV and pursue structure determination, as well as LP3 continued to offer services within biophysical characterization of proteins. LP3 continued the support to the DEMAX platform of ESS and the FragMAX platform of MAX IV and LP3 became associated with the SciLifeLab Lund node as a local research infrastructure.
LP3 staff was in 2023 involved in both undergraduate and graduate teaching, as well as national and international conferences and networks of interest.
In general, both within and outside PPS, LP3 continued in 2023 to deliver projects to its users at the maximum of its capacities.
Wolfgang Knecht,
Manager LP3
April. 2024
Introduction
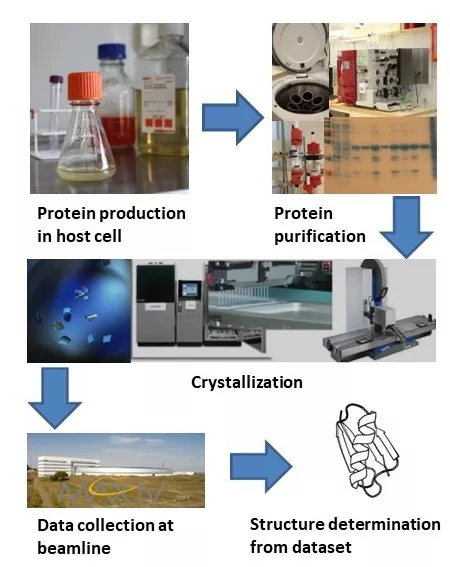
Lund Protein Production Platform (LP3) is a center at Lund University (LU) that offers services and equipment in the areas of recombinant protein production, protein crystallization, biophysical characterization, and structure determination as illustrated in Figure 1. The center was established by the Faculty of Science (FoS), Medicine (FoM) and Faculty of Engineering (LTH) in 2016 (STYR 2023/2145). LP3 serves as the LU node of the distributed national research infrastructure Protein Production Sweden (PPS, external webpage) since 2022 and is a part of the MAX IV laboratory X-ray-aided fragment screening platform (FragMAX, external webpage) that was established by a Swedish research council (VR) grant with LP3 as co-applicant since 2019.
LP3 provides labs and services for and collaborates with the Deuteration and Macromolecular Crystallization platform (DEMAX, external webpage), of the European Spallation Source ERIC (ESS), currently as a Swedish in kind contribution to ESS (see the ESS external webpage and the Swedish Research Council external webpage). The labs managed by LP3 serve as a meeting point for all these platforms. LP3 has staff equivalent to 6 full-time employees. Of this workforce roughly 50 % support the operations of the LU node of PPS (also referred to as LP3-PPS), 10 % support FragMAX, 10 % deliver the Swedish in-kind contributions to ESS and 30 % support crystallization and structure determination operations of LP3, as well as all other local support. Both FragMAX and DEMAX also have their own staff placed in the labs provided by LP3. Those parts outside LP3-PPS are also in summary referred to as LP3-local. Just LP3 means in the following parts LP3-PPS and LP3-local.
The crystallization and structure determination operations of LP3 in LP3-local, as well as the FragMAX platform have been selected to be included in the SciLifeLab platform plans for evaluation during 2024, with the possibility to become a part of the Integrated Structural Biology (ISB) platform from 2025 on. LP3 is already associated as a local research infrastructure with the Lund SciLifeLab node and as a LU infrastructure of national importance, as well as listed in Lucris by LU as such (more information of the LUCRIS homepage). Additionally, LP3 is part of LU thematic collaboration initiative: Pandemics and Alertness.
LP3’s mission (STYR 2023/2145) is to:
- offer open service and support, primarily to researchers at LU, with protein production, characterization and crystallization for their research projects.
- be responsible for a common and open infrastructure for protein production and crystallization, as well as to contribute actively to the interaction of LU with MAX IV, ESS and other relevant major research facilities, networks and initiatives.
- act as Lund University's node for the national research infrastructure PPS
- develop competence and methods in the area of protein sciences.
- serve the surrounding community (e.g. closely located large infrastructures, small biotech etc.).
User access is given on a first come basis and technical feasibility. In 2023, LP3 services were fully subscripted, yet sufficient to meet the demand with limited waiting times for users. Extensive work requests, however filling up all available capacity, are divided into smaller parts or rejected in order not to delay delivery times unproportionally for many other users. In general, at LP3-local, LU researchers and LP3-local’s commitments to FragMAX and DEMAX (ESS) are prioritized while for LP3-PPS this is obviously not the case and in case of lacking capacity at PPS, scientific quality of the project application is accessed for prioritization by the management group of PPS.
Organisation
LP3 was run in 2023 by a manager (60 % FTE, senior lecturer) and 5 research engineers. In 2023, the staffing also included 2 experts (10 %) in microbial protein production and crystallization as well as hosting Dr. Z. Fisher (Head of DEMAX (ESS), assoc. lecturer at LU with 20 %).
4 visitors were associated with LP3 in 2023, not counting the staffing of the FragMAX project or DEMAX staff. Additionally, LP3-local and the wet-labs of FragMAX facility have a lot of users (e.g. in 2023 33 users) coming to LP3’s labs to use certain equipment themselves or doing short experiments at one or several occasions.
LP3 fully equipped for protein production in E. coli and insect cells (Baculovirus Expression Vector System, BEVS). This includes flow hoods for sterile handling of cells, temperature-controlled shakers for culturing of cells (including access to temperature-controlled rooms), centrifuges, cell homogenization equipment (e.g., French Press and sonicators). For purification there are several chromatography systems, including one Äkta Avant and two Äkta Pure Systems and an Äkta Flux for diafiltration. Equipment for SDS PAGE, Western blotting and other standard equipment for protein characterization and enzymatic activity assays is available at the center or within close proximity. All documentation is captured using electronic lab notebooks. For crystallization the facility is equipped with state-of-the-art nanolitre pipetting equipment with the capability to handle lipidic cubic phases for membrane proteins, as well as “plate hotels” with the capacity to store and automatically image plates. Tecan and a TTP (Dragonfly) liquid handling systems for the preparation of crystallization screens are also available. For characterization of proteins LP3 can provide differential scanning fluorimetry (nanoDSF), dynamic light scattering system (DLS) and a size exclusion chromatography (SEC) system (with integrated multi-detector module) for the measurement of absolute molecular weight, molecular size and sample composition (OMNISEC).
Find more specifics on the capabilities and services of LP3 on our homepage.
Placement of the infrastructure
LP3 is placed at the Biology Department (Biology Building A, Sölvegatan 35, 22362 Lund), within the Faculty of Science (FoS) at LU. LP3 is a separate entity within the existing administrative structure of the Department of Biology and follows the working and delegation principles of the FoS.
Leadership of the infrastructure
LP3 is governed by a board of one chairman (Prof. Anders Tunlid) and 6 members (Prof. Susanna Horsefield, Dr. Kajsa Paulsson, Prof. Mats Ohlin, Dr. Kajsa Sigfridsson Clauss, Dr. Sindra Petersson Årsköld), one each from Faculty of Science (FoS), Faculty of Medicine (FoM), LTH, MAX IV laboratory and ESS (external member) and one student (vacant during 2023). The chairman is the dean or vice dean of the FoS. The daily business of the center is led by a manager (60 % FTE) (Dr. Wolfgang Knecht). The manager is supported in his function by additional experts (10 % FTE) (Currently Dr. Claes von Wachenfeldt (microbiological protein production and deputy manager LP3) and Dr. Derek Logan (crystallization).
PPS – Summary from the annual report 2023
Protein Production Sweden (PPS) has in its second year continued its establishment as a national infrastructure. All project applications can now be entered through the common web application form into our project database and are then discussed in the management group and distributed to the different labs. In this second year, 137 PIs applied for 197 different projects, increases corresponding to 30 and 40% respectively, compared to 2022. This shows the continued high demand for recombinant proteins from the Swedish scientific community. During 2023, 168 projects were completed, giving rise to the delivery of 324 protein batches (47 % increase compared to 2022) showing that our infrastructure has gained momentum, but also bringing us to the edge of our capacities. To continue to spread the information about PPS among Swedish researchers, we have presented our capabilities at different conferences and meetings on 24 different occasions. New users have come, but the second year has also seen many returning users from 2022, and PPS received very favorable user survey results again this year. We have agreed upon methods that ensure the quality of the delivered proteins. Moreover, we have worked on several development activities to establish more protocols that our users can now take advantage of.
Details about LP3s part in PPS can be found in the PPS annual report to the Swedish Research Council.
FragMAX (BioMAX Fragment Screening platform)
The FragMAX platform of MAX IV (external webpage) is a Swedish Research Council financed project, with LP3, AstraZeneca AB and Saromics Biostructures AB as co-applicants. The project aims at setting up high throughput X-ray fragment screening (XFS) at the BioMAX beamline with LP3 as the partner for crystallization and sample preparation (for more information please see the publication by Lima et al. in Acta Crystallographica 2020 (external website)). FragMAX has become a permanent platform within MAX IV laboratories with its “wet labs” firmly established at LP3.
Deuteration and Macromolecular Crystallization (DEMAX)
The DEMAX platform of the European Spallation Source ERIC (ESS) is co-localized with LP3 since 2016. DEMAX and LP3 are collaborating to coordinate their efforts to develop cost-effective production of deuterated biomaterials (lipids and proteins) for neutron-based methods such as protein crystallography, neutron reflectometry, and small angle neutron scattering. The underlying agreement for this was renewed for the period 2021-2025.
Dr. Knecht on behalf of LP3 applied within the first VR (Swedish Research Council) call for in-kind contributions to ESS for providing deuteration lab services. This proposal was accepted by VR and ESS. LP3 is thereby among the very first Swedish in-kind contributions to ESS. Feel welcome to read more about this on the ESS homepage, (external webpage) and on the Swedish Research Council homepage (external webpage).
Long term strategy
The full-length strategic plan for LP3 can be found in the LP3 annual report 2021 (internal webpage). In short, LP3 has the ambition within PPS (LP3-PPS) to become and remain a) Preferred supplier of (per)deuterated biomolecules for neutron scattering experiments b) Preferred supplier of specialty protein reagents for X-ray crystallography c) Preferred supplier of protein expression in insect cells with the Baculovirus Expression Vector System (BEVS). This aligns with the overall long-term strategy aim of PPS: “PPS shall become and remain the preferred provider of proteins for research purposes for Swedish researchers from academia, public sector and commercial entities”. Outside of PPS (LP3-local), LP3s strategic plan points towards protein characterization, protein structure determination for non-experts (non-crystallographers) and to support the FragMAX at MAX IV and the DEMAX platform of ESS. This aligns well with the strategic LU goal “The potential of MAX IV and ESS is to be fully exploited”.
Services
LP3 offers services for the entire process chain of production, purification, characterization, protein crystallization and protein structure determination and structure refinement or each individual step in the chain (Figure 1).
For details of current services and updates, please see LP3 homepage (internal).
Users and projects
Some overall user statistics will be reported here. The LP3-PPS part within PPS is described in the annual PPS report to the Swedish Research Council. LP3-local’s activities outside PPS, will be described in more detail in this report below.
Overall
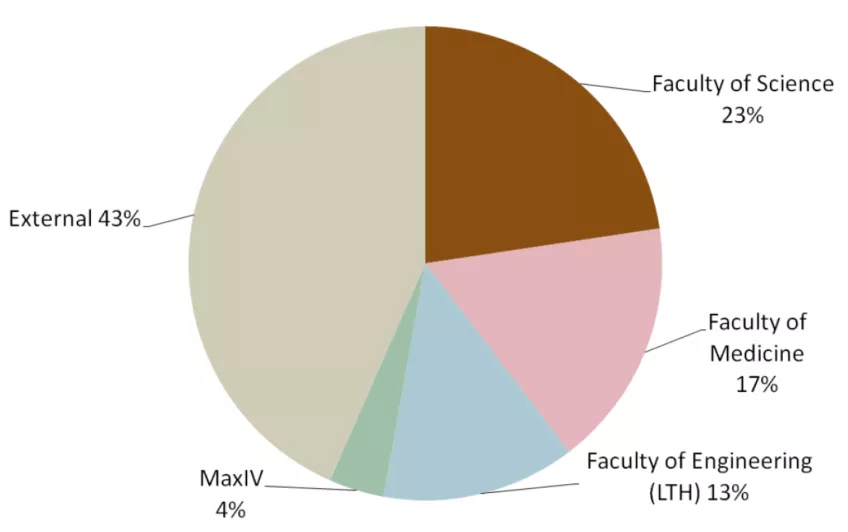
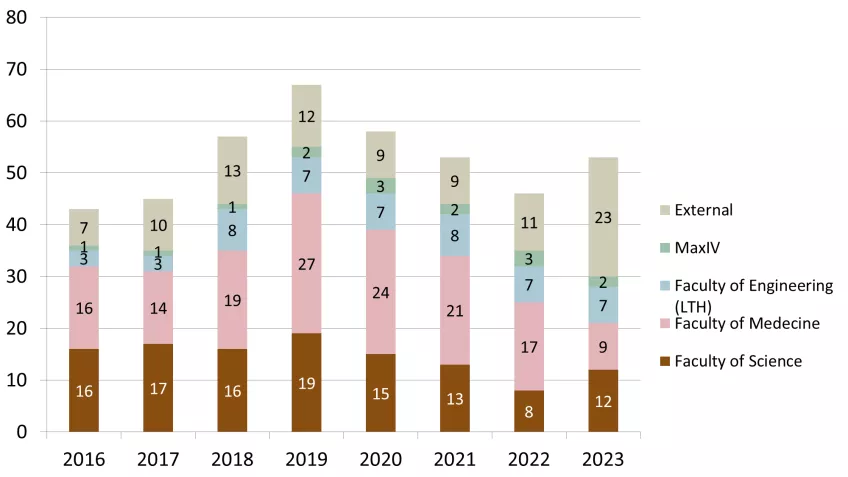
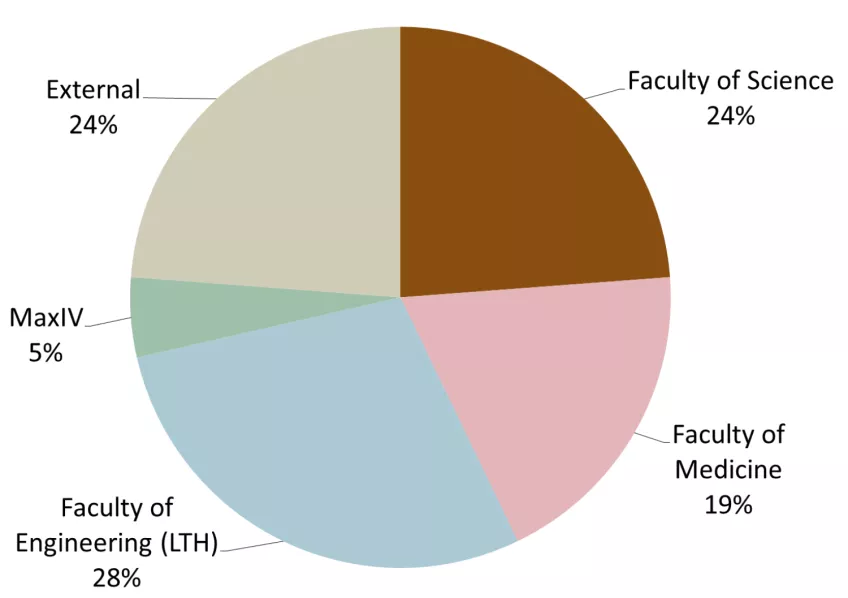
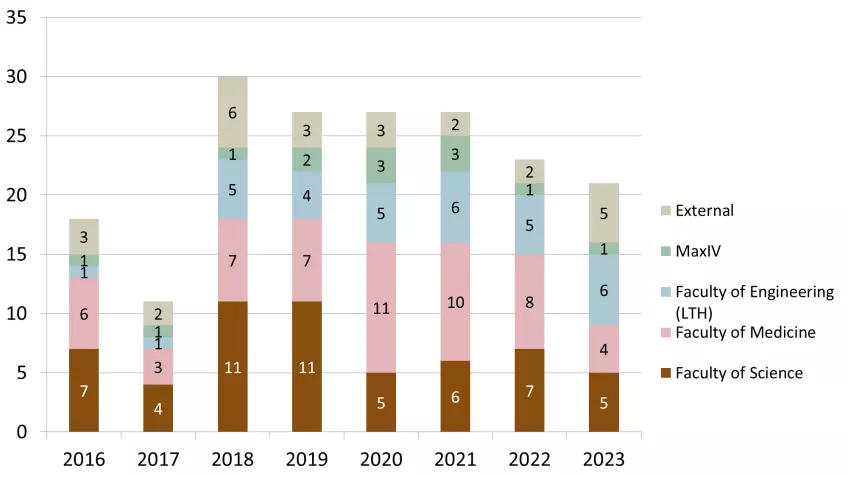
53 groups used LP3 in 2023 both through PPS or as a local infrastructure. Of these, 30 principal investigators came from LU and 23 were external. The distribution to different faculties and external users is presented in Figure 2. The development of user groups in numbers at LP3 since 2016 is shown in Figure 3 and shows the more than doubling of the number of external users of LP3. The principal investigators in the external user group come from the ESS, other Swedish and international universities (for example in 2023: UU, UmU, KTH, LinU University of Oslo (Norway), Amsterdam (Netherlands), Hamburg (Germany), Turku (Estland)) and industry/biotech (in 2023: five companies).
Figure 4 presents the total distribution of 146 unique users at LP3 in the timeframe of 2016 - 2023. Compared to 2022 when this number was 130, this is an increase of 16 unique new users.
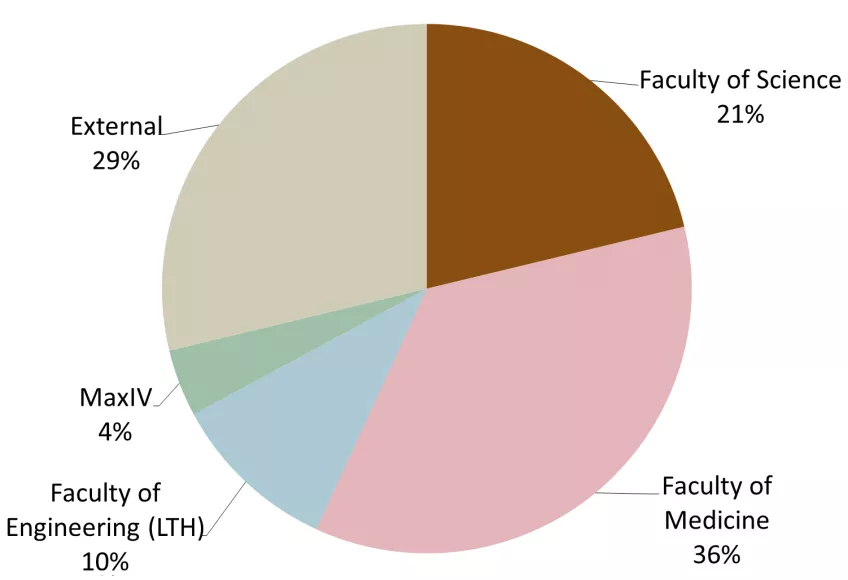
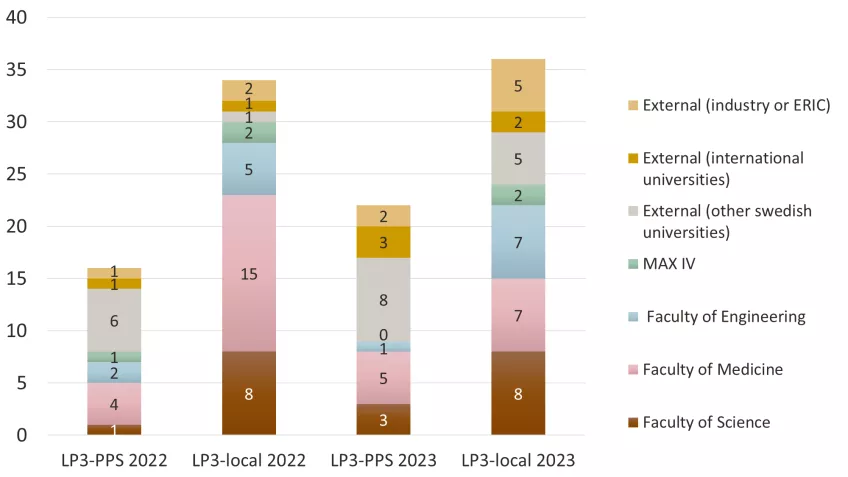
The distribution of users between LP3-PPS and LP3-local (all LP3 activities like crystallization, structure determination, biophysics, local support (e.g. for protein production of previous LP3 projects)) is shown in Figure 5 for 2022 and 2023. Here, the external users are separated into other Swedish universities, international academics and other users (e.g. from industry, public sector and infrastructures like ESS ERIC).
Not included in the numbers above are the “indirect” users that LP3-local served due to supporting all FragMAX campaigns and delivering “Deuteration Lab Services” (DLS) as Swedish in kind contribution to DEMAX (ESS).
LP3-local
In the area crystallization and biophysics, LP3-local, had 21 user groups. A total of 187 screening plates were processed in 2023. LP3-local had 7 projects with the aim of structure X-ray crystallography managed by LP3-local for non-expert users.
The distribution of user groups in crystallization and biophysics and the development of the number of user groups since 2016 are shown in Figure 6 and 7.
8 of the user groups above also used LP3 or PPS for protein production 2023. In 2023, 16 user groups (6 FoS, 2 FoM, 1 LTH, 1 MAX IV, 6 Extern) used other support by LP3-local ranging from production of biomass, production of new batches in previous LP3 projects to technical assistance with Western blotting and production of reagents for undergraduate teaching.
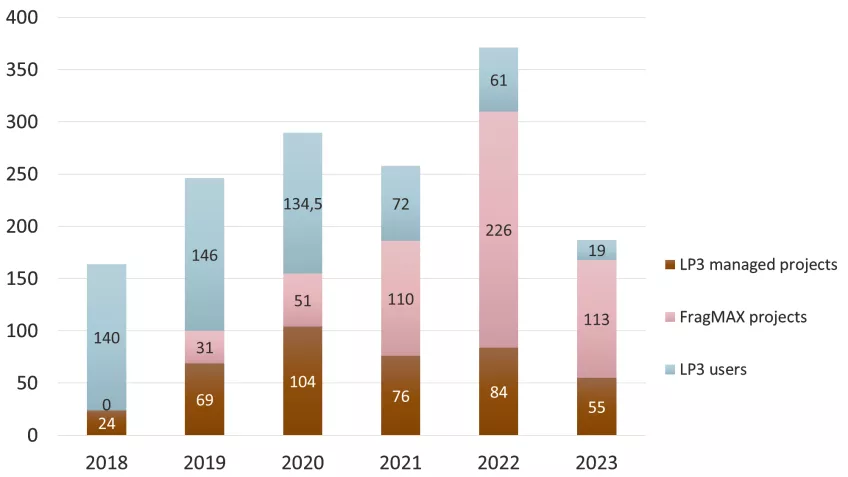
The number of crystallization plates is presented in Figure 8, separated after projects that LP3 runs on behalf of users (non-experts in structural biology), FragMAX projects and projects at which the user sets up the plates themselves.
Crystal screening at the BioMAX Beamline
2023: 318 crystals from 6 projects were tested by LP3 staff at BioMAX on 9 occasions and 101 datasets collected in total. 3 of these projects were managed by LP3 for non-expert users and resulted in 244 crystals and 101 datasets. For the other 3 projects with crystals, LP3 helped expert users to test 74 crystals at the BioMAX beamline.
FragMAX project (BioMAX Fragment Screening platform)
LP3-local supported 12 (8 academic, 4 industry) FragMAX campaigns in 2023. One of those was from LU. Sweden and Scandinavia accounted for 80% of FragMAX users. That is doubling compared to 2022 with 6 campaigns.
As a comment here, we see an increase in numbers from 267 crystals and 70 datasets in 2022 despite less numbers of crystallization plates. Along the same lines, 12 FragMAX campaigns vs 6 in 2023 were supported with less plate set-up.
DEMAX projects
For DEMAX, LP3-local delivered in all 3 areas of the DLS agreed services:
- Development of methods for the production of protiated and deuterated yeast cell paste suitable for lipid extraction.
- Development of production of deuterated biomass and/or proteins and/or DNA.
- Support for neutron protein crystallography
Visibility, access, outreach
LP3 presents its services, capabilities and new developments through Lund University-based homepages (www.lu.se/lp3) and the LUCRIS infrastructure pages, as well as at meetings (see below).
LP3 is featured in EUGLOHRIA Talks and EUGLOHRIA Virtual Tours approaching audiences with interactive and innovative content strengthening the collaboration potential by featuring model core facilities. LP3 was identified as one of those infrastructures available to the Alliance. On the EUGLOHIA Talks homepage (external webpage) an interview with the head of LP3, Dr. Wolfgang Knecht can be found, and it is also possible to take a virtual tour at LP3 on the EUGLOHRIA Virtual Tours external website.
LP3 participates in relevant national and international networks and societies, (e.g., Protein Production and Purification Partnership in Europe (P4EU, external webpage) and Core Technologies for Life Sciences (CTLS, external webpage). Since 2019, LP3 is one of the LU infrastructures affiliated to EATRIS ERIC (the European infrastructure for translational medicine, see external webpage) in their small molecule platform. LP3 is also a member of the Deuteration Network DeuNet (external webpage).
Above, engagements are for dissemination of LP3’s work as well as for the exchange and adoption of new ideas and methods into LP3.
As nearly every year, LP3 was contributing to undergraduate education. At the graduate level, LP3 contributed to the Integrated Structural Biology (SciLifeLab) Ph.D course ”Integrated Structural Biology” and the LU Ph.D course “Protein Factories” with oral lectures about protein production including presentations of LP3 and PPS.
A method protocol of LP3’s standard operating procedures for the BEVS was published – see in publication list below: [10]. Also noteworthy, PPS published a popular science description description “Skördetid i proteinfabriken” (only available in Swedish) in the journal of the Swedish Chemical Society. The article is available at the PPS webpage under news.
Presentations of LP3 and/or of PPS by LP3 staff were provided at the following occasions in 2023:
Oral presentations
LP3 and PPS
- European Conference on Neutron Scattering (ECNS), Munich, Germany, March 2023
- Local Friday seminar of the “Infection research community in Lund”, Lund, March 2023
- LINX: Structure-Based Drug Discovery symposium and course with focus on Fragment Based Lead Discovery, Lund, September 2023
Poster presentations
LP3 and PPS
- Lund Spring Symposium: Enabling Novel Therapeutic Principles, Lund, May 2023
- BSR14 (14th international conference on biology and synchrotron radiation), Lund, June 2023
- 26th Swedish Conference on Macromolecular Structure and Function, Tällberg, June 2023
PPS only
- HALRIC KICK-OFF CONFERENCE, Copenhagen, Denmark, April 2023
- EBSA-2023 Congress, Stockholm, August 2023
Attendance
LP3 and PPS
- P4EU meeting, Trieste, Italy, June 2023
Results and/or proteins produced at the facility were used in the following 2023 publications (in the time 2016 – 2023, the total number of publications has thereby reached 114):
Publications
- Eltschkner, S.; Mellinger, S.; Buus, S.; Nielsen, M.; Paulsson, K. M.; Lindkvist-Petersson, K.; Westerdahl, H. The structure of songbird MHC class I reveals antigen binding that is flexible at the N-terminus and static at the C-terminus. Frontiers in Immunology. 2023, 14. https://doi.org/10.3389/fimmu.2023.1209059
- Frick, I. M.; Happonen, L.; Wrighton, S.; Nordenfelt, P.; Björck, L. IdeS, a secreted proteinase of Streptococcus pyogenes, is bound to a nuclease at the bacterial surface where it inactivates opsonizing IgG antibodies. The Journal of biological chemistry. 2023, 299, 105345. https://doi.org/10.1016/j.jbc.2023.105345
- García-Revilla, J.; Boza-Serrano, A.; Jin, Y.; Vadukul, D. M.; Soldán-Hidalgo, J.; Camprubí-Ferrer, L.; García-Cruzado, M.; Martinsson, I.; Klementieva, O.; Ruiz, R.; Aprile, F. A.; Deierborg, T.; Venero, J. L. Galectin-3 shapes toxic alpha-synuclein strains in Parkinson’s disease. Acta Neuropathol. 2023, 146, 51-75. https://doi.org/10.1007/s00401-023-02585-x
- Happonen, L. J. Affinity-Purification Combined with Crosslinking Mass Spectrometry for Identification and Structural Modeling of Host-Pathogen Protein-Protein Complexes. Methods Mol Biol. 2023, 2674, 181-200. https://doi.org/10.1007/978-1-0716-3243-7_12
- Kettisen, K.; Nyblom, M.; Smeds, E.; Fago, A.; Bülow, L. Structural and oxidative investigation of a recombinant high-yielding fetal hemoglobin mutant. Frontiers in Molecular Biosciences. 2023, 10. https://doi.org/10.3389/fmolb.2023.1133985
- Knecht, W.; Fisher, S. Z.; Lou, J.; Sele, C.; Ma, S.; Rasmussen, A. A.; Pinotsis, N.; Kozielski, F. Oligomeric State of β-Coronavirus Non-Structural Protein 10 Stimulators Studied by Small Angle X-ray Scattering. Int J Mol Sci. 2023, 24. https://doi.org/10.3390/ijms241713649
- Mahanti, M.; Pal, K. B.; Kumar, R.; Schulze, M.; Leffler, H.; Logan, D. T.; Nilsson, U. J. Ligand Sulfur Oxidation State Progressively Alters Galectin-3-Ligand Complex Conformations To Induce Affinity-Influencing Hydrogen Bonds. Journal of Medicinal Chemistry. 2023, 66, 14716-14723. https://doi.org/10.1021/acs.jmedchem.3c01223
- Palica, K.; Deufel, F.; Skagseth, S.; Di Santo Metzler, G. P.; Thoma, J.; Andersson Rasmussen, A.; Valkonen, A.; Sunnerhagen, P.; Leiros, H. S.; Andersson, H.; Erdelyi, M. α-Aminophosphonate inhibitors of metallo-β-lactamases NDM-1 and VIM-2. RSC Med Chem. 2023, 14, 2277-2300. https://doi.org/10.1039/D3MD00286A
- Raum, H. N.; Fisher, S. Z.; Weininger, U. Energetics and dynamics of the proton shuttle of carbonic anhydrase II. Cell Mol Life Sci. 2023, 80, 286. https://doi.org/10.1007/s00018-023-04936-z
- Sullivan, H. M.; Krupinska, E.; Rasmussen, A. A.; Orozco Rodriguez, J. M.; Knecht, W. (2023) Recombinant Protein Production Using the Baculovirus Expression Vector System (BEVS) in Advanced Methods in Structural Biology (Sousa, Â. & Passarinha, L., eds) pp. 55-77, Springer US, New York, NY. https://doi.org/10.1007/978-1-0716-3147-8_4
- Wagner-Egea, P.; Aristizábal-Lanza, L.; Tullberg, C.; Wang, P.; Bernfur, K.; Grey, C.; Zhang, B.; Linares-Pastén, J. A. Marine PET Hydrolase (PET2): Assessment of Terephthalate- and Indole-Based Polyester Depolymerization. Catalysts. 2023, 13, 1234. https://doi.org/10.3390/catal13091234
- Wernersson, S.; Birgersson, S.; Akke, M. Cosolvent Dimethyl Sulfoxide Influences Protein–Ligand Binding Kinetics via Solvent Viscosity Effects: Revealing the Success Rate of Complex Formation Following Diffusive Protein–Ligand Encounter. Biochemistry. 2023, 62, 44-52.
Collaboration
We collaborate in various ways with wider society and organizations within and outside LU. In 2023 LP3 hosted 4 visitors. LP3s efforts for visibility, access and outreach are described in the equally named chapter above. LP3 is part of PPS, which results in collaborations between the different nodes and platforms in PPS. LP3 became associated in late 2023 as a local research infrastructure with the Lund SciLifeLab node. Additionally, LP3 is part of LU thematic collaboration initiative: Pandemics and Alertness (external webpage).